Documentation
Specifying chemical reaction networks - Model properties | Back to contents
In order to specify chemical reaction networks, you need to understand the Model properties panel. In the following, we describe and explain all the details related to the Model properties.
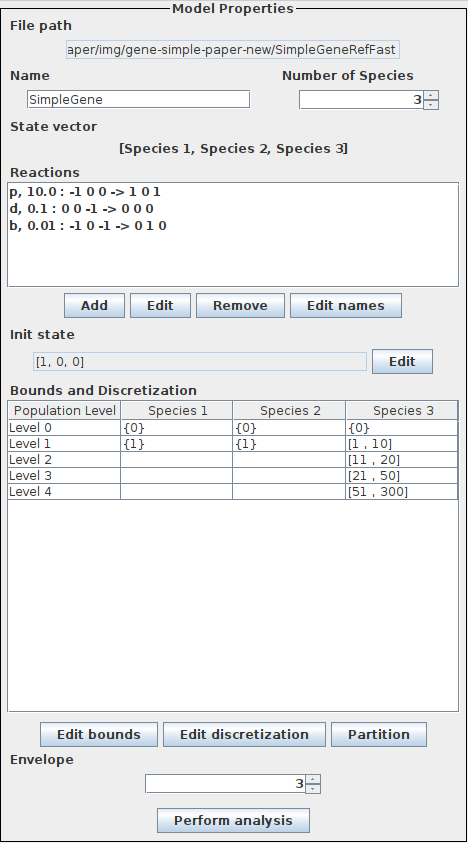
The figure shows the model properties for the simple gene expression model. Refer to the quickstart guide for a shorter description.
File path
Indicates where the model will be saved. If it is empty, no file path is currently specified.
Name
The name of the model. The name may be left empty.
Number of Species
Specifies the number of species in the chemical reaction network. Make sure to decide on the number of species before adding reactions, editing the discretization, editing the bounds and editing the initial state.
State vector
Indicates which component corresponds to which species. In particular, this is important for specifying the reactions and editing the initial state and bounds.
Reactions
Displays a list of reactions. The list items are formatted like this: [Reaction label],[Rate]: [r_1] ... [r_d] -> [p_1] ... [p_d].
d is the number of species in the chemical reaction network, [r_1] ... [r_d] are non-positive stoichiometric coefficients of the reactants and [p_1] ... [p_d] are non-negative stoichiometric coefficients of the products.
Adding reaction: Click Add beneath the list of reactions. A dialog opens, where you can specifiy the Reaction label, stoichiometric coefficients of the reactants, stoichiometric coefficients of the products and rate of the reaction. The first spinner in each line corresponds to the coefficient for the first species, the second for the second species and so fourth. Note that the stoichiometric coefficients of the reactants have to be non-positive and the coefficients of the products have to be non-negative.
Editing reactions: Select a reaction from the reaction list and click Edit below the list. A dialog, similar to the dialog for adding reactions, opens.
Removing reactions: Select a reaction from the reaction list and select Remove beneath the list.
Edit species names: Click Edit names beneath the reaction list. A dialog opens, where you can change the name of the species. Note that you have to click Apply to save the changes. Once applied, the state vector will update and you can see the new name(s).
Init state
Indicates the current initial state, i.e. the initial population for all the species. The components of the initial state correspond to the components of the state vector (see above).
Edit init state: Click Edit. A dialog opens where you can edit the initial population of each species. Make sure you actually enter the population and not the population level.
Bounds and Discretization
Displays the different population levels and the corresponding intervals.
Editing the bounds: Click on Edit bounds. A dialog opens, where you can edit the maximal population each species can attain. By default, the population bound for each species is 2^31 - 1.
Editing the discretization: Click on Edit discretization. A dialog opens, where you can select the species for which you want to edit the discretization. Enter an integer value and click Add to add it to the discretization. The thresholds are automatically sorted. Select a threshold and click Remove to remove the threshold. Suppose the lists consists of thresholds t_1,...,t_l. This corresponds to the population levels [0,0],(0,t_1],(t_1,t_2],...,(t_l,bound].
Partition: Select a population level of a species in the discretization table and click Partition. A dialog opens, where you can specify the number of intervals n the population level should be partitioned into. The population will be partitioned into n equally large intervals.
Envelope
Essentially, the envelope controls how many transitions to consider. If the envelope is set to 0, SeQuaiA will only consider the most probable behavior, i.e. the fastest transitions in each state. Thus, increasing the envelope will incorporate less probable behavior, i.e. slower transitions. The envelope is useful for studying less probable behaviors of the system and for countering non-determinism. By default, the value is set to 4.